Tutorial
DSSR manual
In progress...
1. Analysis
The analysis component of Web DSSR determines a wide range of spatial parameters from a user-provided PDB/NDB ID (the structure identifiers used respectively in the Protein Data Bank and the Nucleic Acid Database) or a PDB-formatted file. The output files can be downloaded or viewed directly on a results page. Selected features and torsion angle parameters are reported in interactive tables.
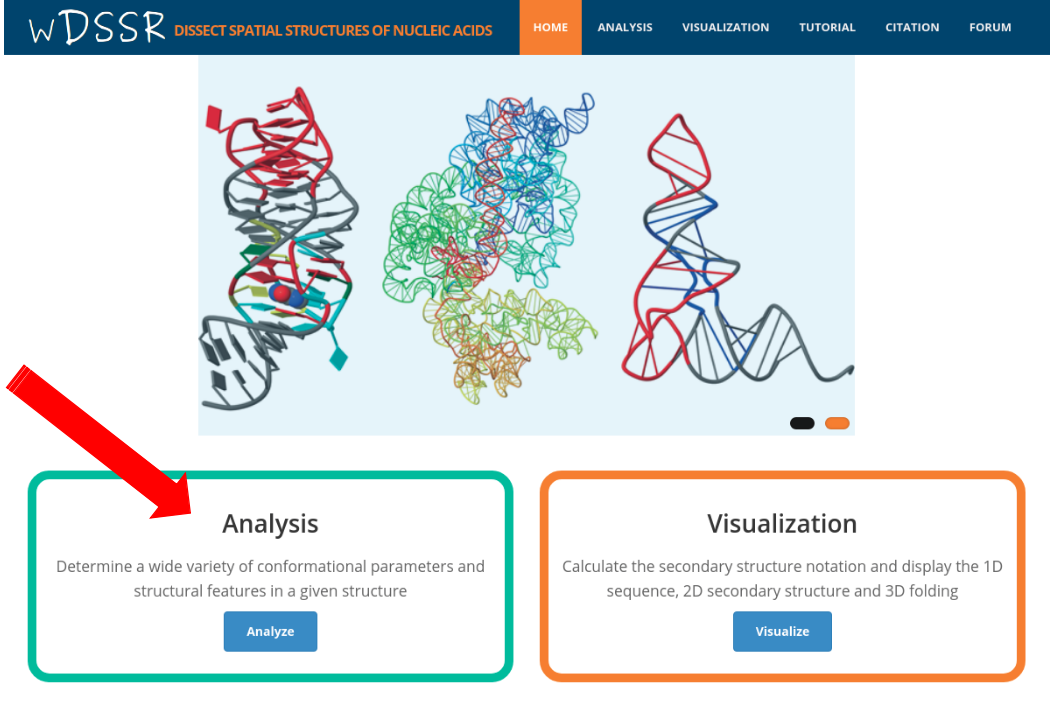
After click the "Analyze" button on the home page, the user is directed to the analysis component. Before conducting an analysis on the web server, users are encouraged to verify the PDB/NDB ID of the input structure at the PDB or NDB website and gain some familiarity with the structure. To use the analysis component, the user can enter a valid PDB/NDB ID in the input area, for example "1EUY". wDSSR can select a biologically active unit according to PDBe annotation.
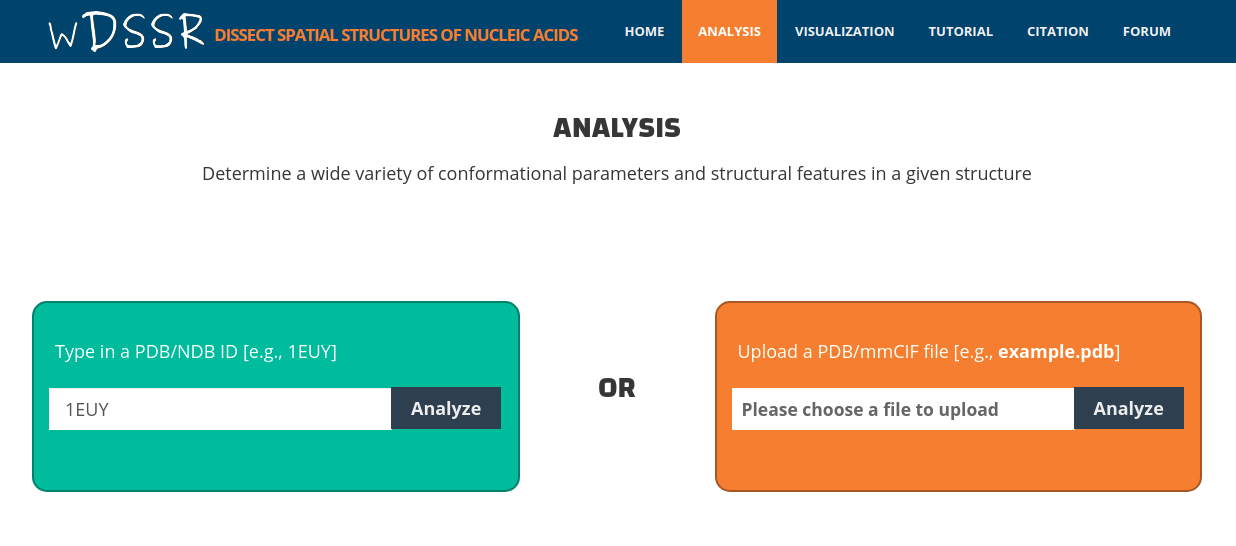
After clicking the 'Analyze' button, the user is directed to a results page containing:
1. Structural summary

2. Interactive tables and 3. Interactive 3D view
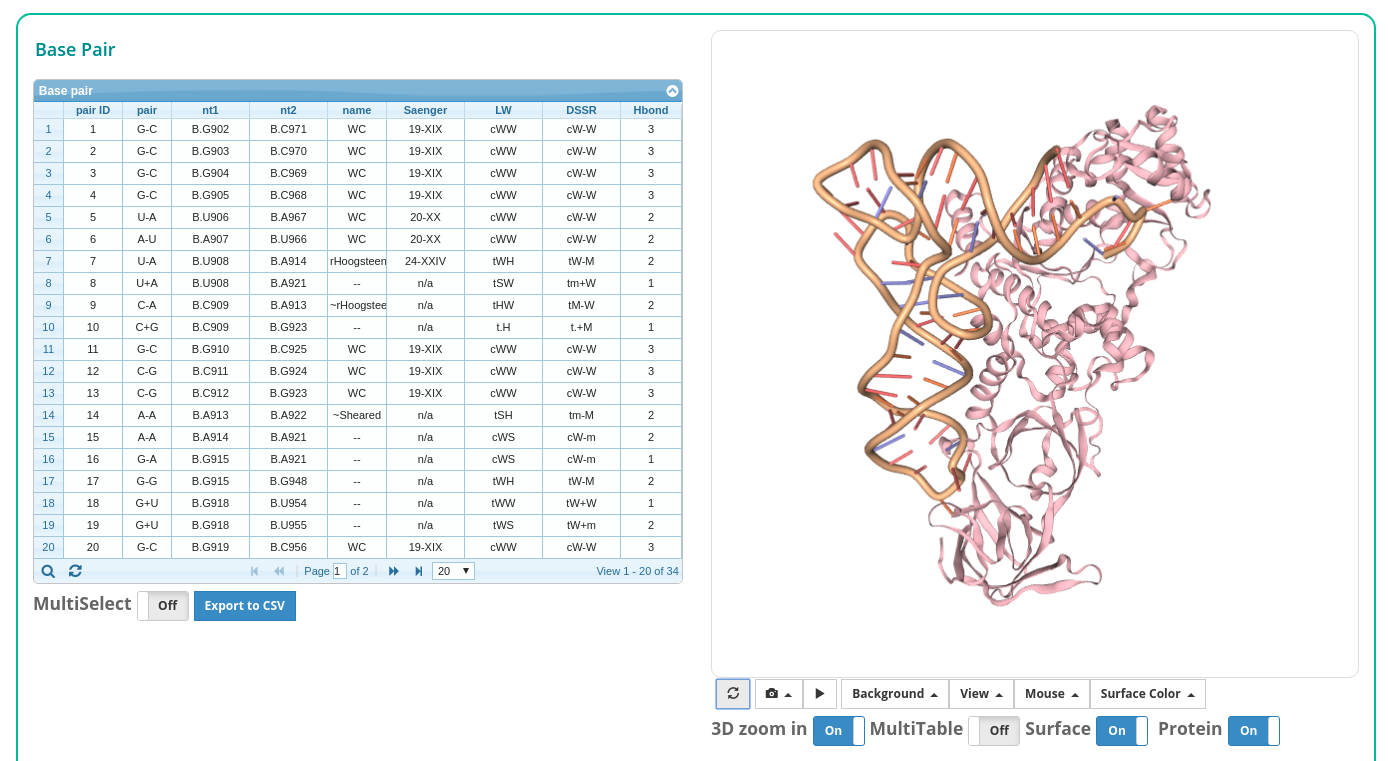
4. A navigation pane
The navigation pane shows all the features found for the input structure. The user can click each feature on the pane to see the details.

2. Visualization
The visualization component of Web DSSR calculates the secondary structure notation and display the 1D sequence, 2D secondary structure and 3D folding
After click the "Visualize" button on the home page, the user is directed to the visualization component. To use the visualization component, the user can enter a valid PDB/NDB ID in the input area, for example "1EUY". wDSSR can select a biologically active unit according to PDBe annotation.
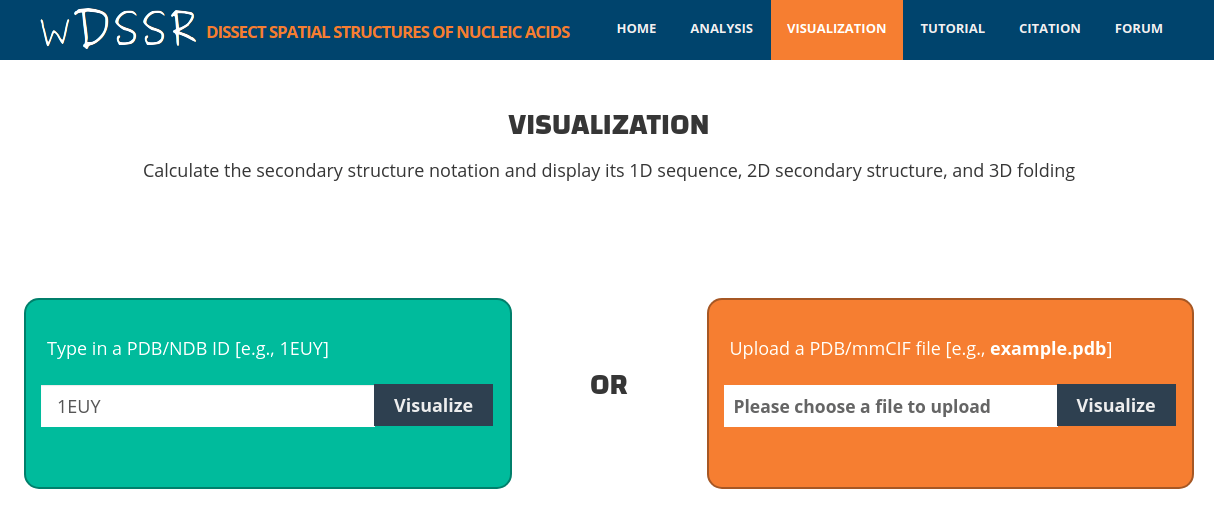
After clicking the 'Visualize' button, the user is directed to a results page containing:
1. 1D sequence control, 2. 2D secondary structure control and 3. Interactive 3D view
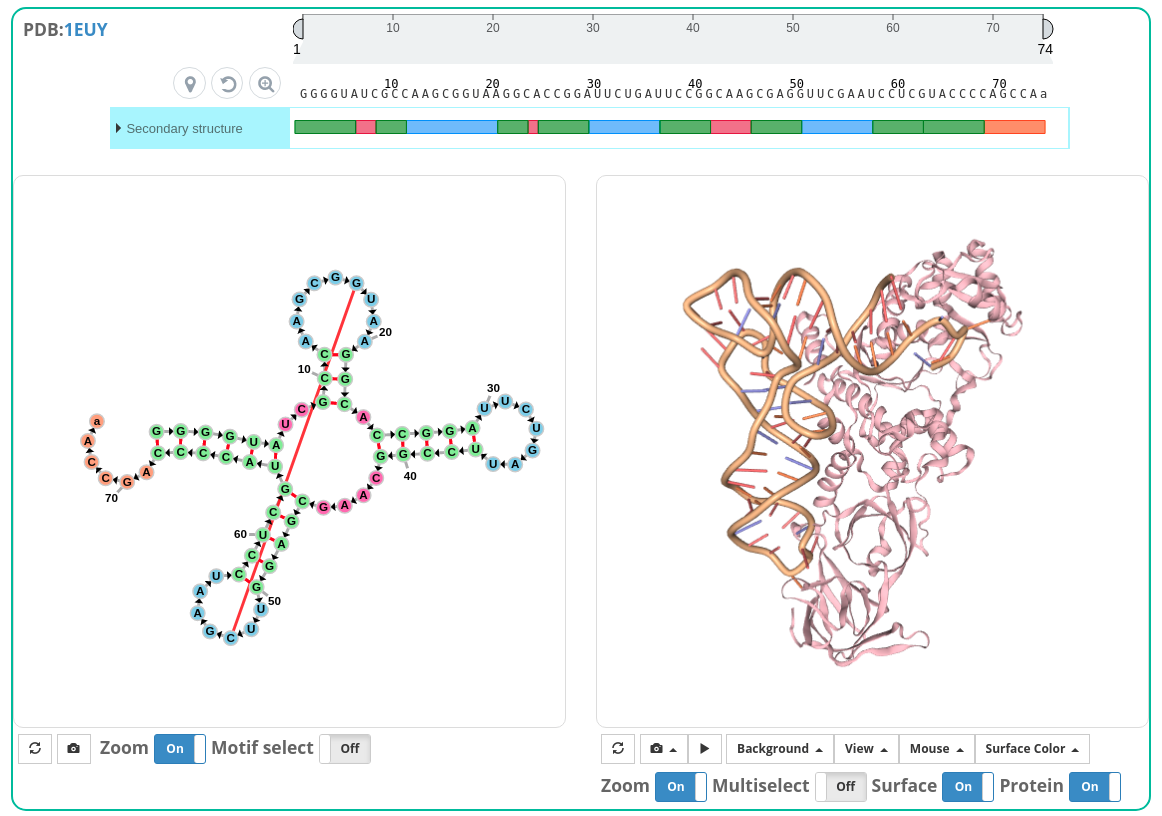
The user can click nucleotide or motif to see simultaneous displaying of selection on each view.
2019 Shuxiang Li@Prof. Wilma K. Olson Group, Rutgers University, New Jersey, USA
Dr. Xiang-Jun Lu, Columbia University, New York, USA
Dr. Xiang-Jun Lu, Columbia University, New York, USA